
Alaleh Azhir was one of 32 American students recently selected from an applicant pool of 880 for a Rhodes Scholarship, which is among the oldest and best-known awards for international study. The link to a JHU press release is https://hub.jhu.edu/2018/11/19/alaleh-azhir-rhodes-scholar
Since the beginning of her sophomore year, Azhir has worked in the lab of Dr. David Nauen, an Assistant Professor of Pathology at the Johns Hopkins School of Medicine. She helped design and build a tool for visualizing and comparing changes in gene expression that the lab uses to investigate the development of epilepsy. Epilepsy is surprisingly common and can have tremendous adverse impacts. In temporal lobe epilepsy, a genetically typical individual suffers an insult such as trauma or fever. Following a prolonged symptom-free period, sometimes many years later, patients start to experience spontaneous seizures. These are centered in the hippocampal formation.
The hippocampus has distinctive circuitry and some of its cells change gene expression rapidly after activity. These properties appear to contribute to its central role in storing memories of experiences. But the same properties likely also make this region prone to epilepsy. To investigate the mechanisms, the Nauen lab is looking at gene expression in individual cells, which lets them assess the changes by the type and developmental state of the cell. Combining this information with knowledge about the stage of epilepsy development, they hope to discover the mechanisms that are quietly altering the hippocampus after that first insult. The ultimate goal is to make the process preventable.
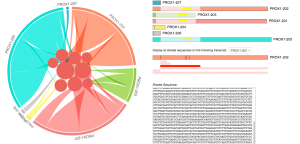
In their experiments, after converting each cell’s mRNA to cDNA, they sometimes use PCR to look at levels of individual genes. Alternative splicing seems to play major roles in neuronal biology, but in designing primers they realized that researchers didn’t know which splice variants might be most expressed in the cells of interest in the various conditions. They also wanted amplicons to cross exon-exon boundaries, they could be sure to look at real mRNA signal rather than contamination from genomic DNA.
(Screenshot from the Trac tool, located at http://labs.pathology.jhu.edu/nauen/trac/)
Alaleh’s work was critical in developing a tool to automatically download splice variants, which are continually being discovered, from public repositories, compare these to find overlapping regions, and display the results in a new way that is interactive and packs a lot of information into an easily understood format. “Besides being powerful and showcasing her creativity, it is potentially of value to anyone who wants to characterize gene expression by PCR or in situ hybridization,” Dr. Nauen said. ”She’s been an amazing student to work with, and I am proud to think that maybe in a small way I’ve helped shape the trajectory of someone with so much promise .”