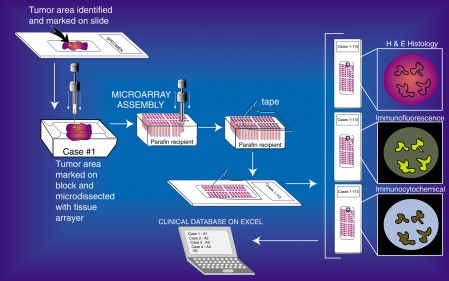
Tissue Microarray Process (click for larger)
Tissue Microarrays (TMAs) have been validated as useful tools for the evaluation of various biomarkers, especially in oncology specimens. To construct a TMA, areas of interest are marked on the hematoxilyn & eosin (H & E) slides. Next, the corresponding area on the matching paraffin block (donor block) is then identified and marked. Tissue cores are removed from these areas of the donor blocks, one by one and inserted into the recipient block in a precisely arrayed fashion. The resultant TMA may contain up to 1,000 individual tissue cores. These miniaturized collections of tissue spots result in a dramatic increase in throughput for in situ examination of gene status and gene expression from archival specimens. All standard methods for examination of standard histological sections, including immunohistochemistry, in situ hybridization, DNA ploidy analysis, nuclear morphometry, and fluorescent in situ hybridization can be performed on sections from TMA blocks. Since many clinical specimens contain limited tissues for study, TMAs can expand the number of studies that can be performed from these extremely valuable specimens. The number of slides that can be generated from the TMA block varies according to the depth of the tissue in the original paraffin blocks which, of course, varies. A TMA block generated from donor blocks that have adequate tissue (e.g., ~2-3 mm of tissue in depth) can yield a few hundred 4-5 μm sections.
We have found that very useful arrays can be constructed from an assortment of normal human tissues obtained as discarded material from surgical specimens. These arrays are ideal for working up new antibodies or new probes for in situ hybridization. A second type of array is one with a small number of disease tissues from several specimens as well as adjacent normal tissues. An array with 20 cases of disease and normal tissues can be used to quickly assess a new marker. Many other types of TMAs can be made, as well. For example, one can design TMAs from patients in clinical trials who have been treated with different agents, or one can design a “case-control” series in which some patients progressed after treatment and others did not. These types of arrays typically have materials from many hundreds of patients, sometimes with patient materials obtained from multiple institutions. Then biomarkers are evaluated on the TMAs to determine which markers can predict response to therapy or clinical outcome. Another example is that cell lines and/or xenografts, with known or unknown biomarker/genetic status, can be arrayed. The JHU TMA lab has so far constructed 620 TMA blocks, with 102,135 cores from 11,978 cases for various JHU researchers.
This is an image of 200 separate TMAs laid out side-by-side that were generated in the JHU TMA lab for various investigators. This shows the range of type of TMA blocks that can be produced using several different core diameters and different types of design.
For handling TMA data, Johns Hopkins researchers have developed open source software (TMAJ) that is being used extensively at JHU and at a number of other institutions, which securely houses data on specimens, TMA blocks, and biomarker data. Other solutions exist, including managing your data simply in a spreadsheet.
TMA slides that have been stained for biomarkers of interest can be examined directly under a microscope, or can be scanned using various whole slide scanners such as the Aperio ScanScope CS. More information about scanning can be found here. Scanned images can also be used for automated and semi-automated quantitative image analysis.
Some of the advantages of using Tissue Micro arrays are:
- High throughput
- Expands tissue use
- Uniform reaction conditions
- Built-in controls
- Economize use of reagents
- Facilitates data recording and linking to clinical data
For more information: http://tmalab.jhmi.edu/tma_construction.html
Angelo De Marzo, M.D., Ph.D.
Professor of Pathology and Oncology
Helen Fedor
Tissue Microarray Lab Manager
